Maghreb/Berber Genetics! ✅
Hi friends, so here is the first official post in the archive. It will be dealing with all the relevant material on the ancient and modern peoples of the Maghreb, enjoy this great collection of compiled data, and happy reading!
List of Ancient DNA Findings (and Anthropometrics):
Iberomaurusian (Taforalt/Afalou) (~15,000 KYA) (🧬N=68)
🔗Open Research: The terminal Pleistocence and early Holocene populations of northern Africa (anu.edu.au)
Based on dental traits in 2000, Joel D. Irish found a relationship between the Iberomaurusians, particularly those from Taforalt, and later Maghreb and other North African samples. Thus, some measure of long-term population continuity in the Maghreb and surrounding region is supported, whereas greater North African population heterogeneity during the Late Pleistocene is implied with strong differences with Jebel Sahaba, but similarities between Taforalt and Afalou.
🔗(PDF) The Iberomaurusian Enigma: North African Progenitor or Dead End? (researchgate.net)
In 2005, an older study extracted mtDNA from 23 remains from Taforalt, derived from various skeletal material. Mitochondrial diversity in the analyzed samples shows the absence of Sub-Saharan African haplogroups suggesting that Iberomaurusian individuals had not originated in the Sub-Saharan region. The results had speculated a probable local origin and evolution of this population, and confirmed genetic continuity in North Africa based on the continued presence of these markers. It revealed 13 haplotype subclades from the main, H, U, JT, V, U6.
🔗Diversité mitochondriale de la population de taforalt (12.000 ans BP - Maroc): Une approche génétique à l'étude du peuplement de l'Afriquedu nord - Kefi - 2005 - ANTHROPOLOGIE (mzm.cz)
Kefi, Rym et al. (2018), found most of the mtDNA genetic structure of TAF and AFA specimens to contain only North African and Eurasian maternal lineages. These finding demonstrate the presence of these haplotypes in North Africa from at least 20,000 YBP. The very low contribution of a Sub-Saharan African haplotype in the Iberomaurusian samples was also confirmed.
🔗On the origin of Iberomaurusians: new data based on ancient mitochondrial DNA and phylogenetic analysis of Afalou and Taforalt populations - PubMed (nih.gov)
🔗Whole mitogenomes reveal that NW Africa has acted both as a source and a destination for multiple human movements. - Abstract - Europe PMC
 |
| PCS Calculations from Present-Day Populations of Africa, Near-East and Southern Europe |
 |
| *credit:🔗Ancestral Whispers |
🔗Pigmentation of Iberomaurusians | Genetiker (wordpress.com)

A 2019 study seeking to determine if North Africans descend from strictly Palaeolithic groups (Taforalt), or subsequent migrations, discovered that most of the genetic variation in the region was shaped during the Neolithic phases. While the ancient samples had more of the Taforalt component, it is most frequent today in Western North Africans (Saharawi, Moroccans, Algerians) and Berbers, and suggested a continuity of this autochthonous North African component. The consideration of Berber-speaking groups as the autochthonous peoples of North Africa was reinforced by these results, and the ~5th Century BCE Canary Guanches, also clustered with current North Africans (in agreement with their Berber origins). Medieval geneflow events, such as the Arab expansions also left traces in various African populations, but with Neolithization having a much larger demographic impact than the Arab expansions. The results using PCA, Admixture, and internal f3 tests, ultimately showed that the North African genetic landscape was shaped during prehistory. A review by Prendergast et al. (2022), similarly concluded that: “Genome-wide aDNA data from sites in Morocco highlight genetic transformations associated with Neolithization, demonstrating an influx of Eurasian ancestry attributed to trans-Gibraltar population movements between the Early and Late Neolithic periods (~7 ka to ~5 ka). Combining ancient and present-day DNA data, this Neolithic demographic transformation was shown to have a greater genetic impact on present-day North Africans than later processes such as Arabization.”
🔴Khef el Baroud (~5000 KYA) (🧬N=8)
🔗Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe - PMC (nih.gov)
 |
| *credit:🔗Expansions of Anatolian Farmers into Europe and the Maghreb |
🔗https://uu.diva-portal.org/smash/get/diva2:1456257/FULLTEXT01.pdf
🔴Guanches (~2000 KYA) (🧬N=246)
In 1925, based on previous anthropological work an his own studies, that the settlement of the Canary Islands archipelago involved migrants in the late Neolithic, of the Mediterranean race with some Negroid admixture. The Guanche proper were migrants from the mainland closely related to the Berbers of North Africa, who also brought with them sheep and goats.
🔗The Ancient Inhabitants of the Canary Islands - Earnest Albert Hooton - Google Books
The distribution of hair color (from 43 samples), showed no significant difference from today's Canary Island population, while the eye colors of six individuals that could be determined were dark. Dietary analysis indicated a 50-70% reliance on meat and dairy. Overall, the remains had marked sexual dimorphism, tall stature, facial robusticity, which all implied the Guanches were well adapted to the island's environment.
https://www.cambridge.org/core/books/abs/mummies-disease-and-ancient-cultures/mummies-from-italy-north-africa-and-the-canary-islands/A9CE0C6F118A74CE9B6D95F8EC602669
🔗Demographic history of Canary Islands male gene-pool: replacement of native lineages by European - PMC (nih.gov)
🔗The maternal aborigine colonization of La Palma (Canary Islands) | European Journal of Human Genetics (nature.com)
Fregel et al. (2015) examined the mtDNA of Guanches of La Gomera, the results showed that 65% of the examined Gomeros were found to be carriers of the maternal haplogroup U6b1a. The Gomero appeared to be descended from the earliest wave of settlers to the Canary Islands. The maternal haplogroups T2c1 and U6c1 may have been introduced in a second wave of colonization affecting the other islands. It was noted that 44% of modern La Gomerans carry U6b1a, and the La Gomerans have the highest amount of Guanche ancestry among all modern Canary Islanders.
🔗Isolation and prominent aboriginal maternal legacy in the present-day population of La Gomera (Canary Islands) - PMC (nih.gov)
An examination by Ordonez et al. (2017) studied the remains of a large number of Guanches of El Hierro buried at Punta Azul. The 16 samples of Y-DNA extracted belonged to the paternal haplogroups E1a (1), E1b1b1a1 (7) and R1b1a2 (7). All the extracted samples of mtDNA belonged to the maternal haplogroup H1-1626. The Bimbache were identified as descendants of the first wave of Guanche settlers on the Canary Islands, as they lacked the paternal and maternal lineages identified with the hypothetical second wave. In the same year, Rodrigues-Varela et al. (2017) sampled the Y-DNA, mtDNA, auDNA of 11 Guanches buried at Grand Canaria and Tenerife. The 3 samples of Y-DNA extracted all belonged to the paternal haplogroup E1b1b1b1a1 (E-M183), while the 11 samples of mtDNA extracted belonged to the maternal haplogroups H1cf, H2a, L3b1a (3) T2c12, U6b1a (3), J1c3 and U6b. It was determined that the examined Guanches were genetically similar between the 7th and 11th centuries AD, and that they displayed closest genetic affinity to modern North Africans. The evidence supported the notion that the Guanches were descended from a Berber-like population who had migrated from mainland North Africa. Among modern populations, Guanches were also found to be genetically similar to modern Sardinians, as they were determined to be carriers of Anatolian Farmer (EEF) ancestry like contemporary Maghrebis, which probably spread into North Africa from Iberia during the Neolithic. One Guanche was also found to have ancestry related to European Hunter-Gatherers, providing further evidence of prehistoric gene flow from Europe starting 7,000 years ago. In all studies, these aboriginal inhabitants of the Canary Islands were genetically most similar to modern North Africans (Maghrebis) and Berbers. Phenotypically, their Guanche samples with enough SNP coverage were shown to have light and medium skin, dark hair and brown eyes.
🔗Mitogenomes illuminate the origin and migration patterns of the indigenous people of the Canary Islands | PLOS ONE
🔗https://brill.com/display/book/9789004500228/BP000019.xml
 |
| *credit:🔗PCA Showing Present-Day North Africans Clustering with Ancient Guanches and Late Neolithic KEB Moroccans |
Other Findings!
🔗On three skulls from Mechta‐el‐Arbi, Algeria. A reexamination of Cole's adult series - Briggs - 1950 - American Journal of Physical Anthropology - Wiley Online Library
🔗The genomic history of the Iberian Peninsula over the past 8000 years - PubMed (nih.gov)
 |
| Genome Wide Admixture Analysis for Islamic Period Iberians |
🔗Muslims in medieval Europe (angelfire.com)
🔗Hadith Bayad wa Riyad - 13th-century Arabic Love Story – Joy of Museums Virtual Tours
🔗Hadith Bayad wa Riyad, the story of Bayad and Riyadh, 13th century (Qissat Bayad wa Riyad), manuscript Vaticano arabo 368 (riserva) (warfare.gq)
🔗Mural paintings of the Conquest of Majorca | Museu Nacional d'Art de Catalunya
🔗Sala de los Reyes - Nomads Travel Guide (nomads-travel-guide.com)
In 2024, mtDNA and Y-DNA of Islamic burials in Portugal supported a North African origin.
🔗https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0299958
A study in 2021 analyzed the genome for an individual who was buried in the Islamic necropolis of Plaza del Almudín in the city of Segorbe (province of Castellón, Comunidad Valenciana, Spain). He was referred to as “the Giant” by the archaeologists responsible for the excavation, due to his unusual height (184-190 cm) compared with the other individuals found in the site. He was discovered to be of Berber origin, and his uniparental lineages pointed to North African ancestry, but at the autosomal level he displays both North African and European-related ancestries. The individual harbored the Y-chromosome haplogroup E-M81 and mtDNA U6a1a1a.
In 2023, a study examined 9 new sequenced genomes from ancient Moroccan sites, while incorporating older sampled material from the region, in order to fill in archeological and chronological gaps within the Maghreb. The newer autosomal DNA analysis involved; 1 Late Epipaleolithic from Ifri Oberrid (OUB), 5 Early Neolithic from IAM (1) and KTG (4) (Kaf Taht el-Ghar), and 3 Middle Neolithic. The 3 genomes from the Middle Neolithic site in Morocco of Shirat Rouazi (SKH), were modelled as 76.4% Levant Neolithic and 23.6% local Taforalt related ancestry. These SKH migrants from the Near-East were also associated as being the exact same ancestral population that spread goat/sheep pastoralism and contributed to the West Asian influence in the modern populations of East Africa (Savannah Pastoral Neolithic). The KTG populations on the other hand were overwhelmingly of Anatolian Farmer ancestry (EEF). The intensive sequencing of these genomes proved that the Neolithic transitions in North Africa were initiated by migrants. It also showed distinct groups existed in the region initially without demic diffusion, until inter-group admixture occurred later, which is seen in the KEB population. The autochthonous North Africans would transition from hunter-gatherer strategies to adopting farming from these EEF migrants, and herding from the Levantine Neolithic shepards.
 |
| Location of Sampled Genomes & PCA Clustering Diagram |
Contemporary DNA Studies:
Y-Chromosome Data
An older study by Nebel, Almut et al. (2002) concluded that the J-M267 chromosome pool in the Maghreb is derived not only from early Neolithic dispersions, but also from recent expansions of Arab tribes to the Maghreb.
🔗Genetic Evidence for the Expansion of Arabian Tribes into the Southern Levant and North Africa - PMC (nih.gov)
A more thorough study by Arredi et al. (2004), which analyzed populations from Algeria, Tunisia, and Egypt, concludes that the North African pattern of Y-chromosomal variation (including both J1 and E1b1b main haplogroups) is largely of Neolithic origin, which suggests that the Neolithic transition in this part of the world was accompanied by demic diffusion of Proto-Afro-Asiatic or Berber-speaking pastoralists from the Middle East according to Semino, Orella et al. (2004). Although later papers such as Myles, Sean et al. (2005) have suggested that this date could have been as long as ten thousand years ago, with the transition from the Oranian to the Capsian culture in North Africa.
🔗A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa - PMC (nih.gov)
🔗Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area - PMC (nih.gov)
🔗Genetic evidence in support of a shared Eurasian-North African dairying origin - PubMed (nih.gov)
In 2009, 514 samples of various Berber tribes were collected. The results revealed that Mzab Berbers from Algeria had 89.6% E-M215, 1.5% G, 1.5% J and 3.0% R1b. Siwa Oasis Berbers had 28% B, 12% E-M215, 3.2% G, 14% J, 28% R1b. Asni Berbers in Morocco had 85.2% E-M215, Bouhria Berbers 79.1% E-M215, and Middle Atlas Berbers having 81.1% E-M215.
🔗Genetic and linguistic diversities: The Berber and the Berbers (benjamins.com)
Gérard, Nathalie et al. (2009) discovered that there was evidence for the influence of Berber and Arabian DNA from the Moorish occupation found in Southern Iberia. Found again in 2015, associated with E-M81 https://www.sciencedirect.com/science/article/abs/pii/S0378111915001900
🔗Project MUSE - North African Berber and Arab Influences in the Western Mediterranean Revealed by Y-Chromosome DNA Haplotypes (jhu.edu)
 |
| *credit:🔗Geographic Distribution of Y-Chromosome Haplotypes of Select Africam, Middle Eastern, and European Populations |
🔗A revised root for the human Y chromosome differentiation and diversity landscape among North African populations - MedCrave online
https://spiral.imperial.ac.uk/bitstream/10044/1/64507/2/JIG-05-00075.pdf
According to Elkamel, Sarra et al. (2021): "Previous works with Y-chromosome markers have shown that in North Africa the most widespread and common Y lineages are E-M81 and J-M267. E-M81 reaches an average frequency of 45% across the regions (from the countries of Morocco to Egypt) and is thought to have a very recent origin possibly in Northwest Africa. J-M267 is the second most-frequent haplogroup, accounting for around 30% of North Africans and assumed to have spread out of the Arabian Peninsula into North Africa, as is shown by, for instance, its east–west decreasing prevalence. There is wide agreement about the sources of J-M267 in North Africa, which imply multiple and temporarily different expansions of Middle-Eastern and Arabic populations through the Mediterranean. In fact, although this haplogroup is considered to be one of the signatures of the spread of Islam from the Arabian Peninsula, it also retains clues on a much earlier expansion during Neolithic times as part of the previously mentioned Capsian cultural complex that was introduced in North Africa along with agriculture." The researchers also noted that, "considering Tunisian populations as a whole, the majority part of their paternal haplogroups are of autochthonous Berber origin (71.67%), which co-exists with others assumedly from the Middle East (18.35%) and to a lesser extent from Sub-Saharan Africa (5.2%), Europe (3.45%) and Asia (1.33%)."
🔗Insights into the Middle Eastern paternal genetic pool in Tunisia: high prevalence of T-M70 haplogroup in an Arab population - PMC (nih.gov)
Mitochondrial Data
According to the most recent and thorough study on Berber mtDNA from Coudray et al. (2008), which analysed 614 individuals from 10 different regions (Morocco (Asni, Bouhria, Figuig, Souss), Algeria (Mozabites), Tunisia (Chenini-Douiret, Sened, Matmata, Jerba) and Egypt (Siwa)), the results may be summarized as follows: Total West Eurasian lineages (H, HV, R0, J, M, T, U, K, N1, N2, X) at 80% and the total African lineages (L0, L1, L2, L3, L4, L5) at 20%. The Berber mitochondrial pool is characterized by an overall high frequency of Western Eurasian haplogroups, a markedly lower frequency of Sub-Saharan L lineages, and a significant (but differential) presence of North African haplogroups U6 and M1.
🔗The Complex and Diversified Mitochondrial Gene Pool of Berber Populations - Coudray - 2009 - Annals of Human Genetics - Wiley Online Library
There is a degree of dispute about when and how the Sub-Saharan African derived L haplogroups entered the North African gene pool. Some papers suggest that the distribution of the main L haplogroups in North Africa was mainly due to the Islamic era Trans Saharan Slave Trade, as espoused by Harich et al. (2010). Sub-Saharan people did not leave traces in the North African maternal gene pool for the time of its settlement, some 40,000 years ago.
🔗The trans-Saharan slave trade - clues from interpolation analyses and high-resolution characterization of mitochondrial DNA lineages - PMC (nih.gov)
However, in the same year, a study of Berber mtDNA in Tunisia by Sabeh Frigi concluded that some of the L haplogroups were much older and likely introduced from ancient African gene flow around 20,000 years ago. Similarly, El Moncer, Wifak et al. (2010) stated: "All these data considered together suggest that the sub-Saharan component found in Tunisia is rather ancient and could be traced back to the first stage of Neolithic Age (around 9000 YBP), characterized by an ethnic contribution from present-day Sudan." These finding were restated in 2016, with this uniparental evidence pointing to events as far back as 30,000 YBP for certain Sub-Saharan maternal clades. They also noted that: "The Tunisian samples cluster together with the Berber groups from Morocco and Algeria, in agreement with recent works based on other genetic data. The close genetic relationship of the two Arab-speaking populations with the Berber-speaking samples could be explained assuming a small number of Arabs coming from the Arabian Peninsula, as compared with that of the autochthonous Berbers, resulting in a weak Arab genetic influence in the current mixed North Africans." & "In conclusion, the results discussed here allow us to postulate that the general ancient genetic profile of the native North Africans-the Berbers-is not very different from that of the present-day North African populations, despite some admixture with other peoples, particularly Arabs, during successive historical periods. The populations of the Maghreb seem to share a substantial genetic background, regardless of culture and geography."
🔗Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations - PubMed (nih.gov)
🔗Mixed origin of the current Tunisian population from the analysis of Alu and Alu/STR compound systems | Journal of Human Genetics (nature.com)
🔗Genetic variation in Tunisia in the context of human diversity worldwide - PMC (nih.gov)
🔗https://www.academia.edu/4137738/Mitochondrial_DNA_and_Y_chromosome_microstructure_in_Tunisia
In 2013, the haplogroup L3e5 in North African populations was considered to have possibly arrived during the Green Sahara, dated to ~10 kya. They also referenced contact between Northern and Sub-Saharan Africa by the craniofacial data from Niger, Mali and Mauritania, of ancient populations who had Mechtoid affinities like the Iberomarusians and Capsians of the Maghreb. It was taken as evidence for early trans-saharan contact between the two regions.
🔗The genetic impact of the lake chad basin population in North Africa as documented by mitochondrial diversity and internal variation of the L3e5 haplogroup - PubMed (nih.gov)
A study in 2023 that sampled modern haplogroups from 264 Algerian Berbers (Chaoui), found many to be linked to the Neolithization of North Africa from Iberia. The times of coalescence for these clades were ~6,000 ybp for H1cb1, U3a1c, and U5b1b1e. The other European origin lineages like H1e1a, were given a time of ~3000 ybp, as they were also found in the Canary Islands. Middle Eastern origin lineages like J2a2d were dated to ~9,000 ybp, and was said to represent potential pre-agricultural contact between the Near-East and North Africa. T1a7 was associated with Arabization and Bedouin expansion from 1,000 ybp. Sub-Saharan lineages like L3e5, L3b1a9a were dated to ~ 10,000–5000 ybp, and may be from trans-Saharan migration during the Green Sahara period with a prehistoric origin. However, the great majority of sub-Saharan sequences in North Africa do not form phylogenetic clades, and therefore, show no signals of having evolved in North Africa. This is in accordance with a recent arrival for the majority to the region as suggested by the genome wide evidence.
🔗Whole mitogenomes reveal that NW Africa has acted both as a source and a destination for multiple human movements | Scientific Reports (nature.com)
According to Barbujani, G et al. (1994): “Historical data on three comparatively recent episodes of language replacement (the spread of Indo European languages in Asia, of the Turkic subgroup of Altaic languages in Western Asia, and of Arabic in North Africa) suggest that they did not entail major changes in the biological buildup of the populations involved. These were not cases of demic diffusion, but of elite dominance, associated with gene flow only to a very limited extent.”
Based on classical genetic markers, according to Cavalli-Sforza LL, Menozzi P, Piazza A. (1994), the Tuareg have genetic affinities with the Beja people. The inferred ethnogenesis of the Tuareg people happened within a time period of 9,000 to 3,000 years ago, and most likely took place somewhere in Northern Africa. These older findings were loosely referenced in another study from 2010 which also tied the ethnogenesis of the Tuareg with the Garamantes, but said the mtDNA diversity did not indicate an affiliation with the Beja, but instead more ties to Iberian Peninsular. Though they noted future studies on the mitochondrial profile of the Beja tribes of Eastern Sudan will help further clarify these disreprancies.
🔗The History and Geography of Human Genes - Luigi Luca Cavalli-Sforza, Paolo Menozzi, Alberto Piazza - Google Books
🔗Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel - PMC (nih.gov)
From the years of 2005 to 2019, the National Geographic Genographic Project collected samples from populations from around the world into a large genetic database and used various clustering analysis to measure ancestral links. According to the second autosomal runs by National Geographic, the Geno 2.0 Next Generations, the Tunisian population was 88% Northern African, 5% Southern European, 4% Southwest Asia & Persian Gulf, and 2% Central & Western African. In the first Geno 2.0 which instead matched collective ancestry based on regions instead of Biogeographic ancestry like the second which was the final project, the results revealed Tunisians were 62% Mediterranean, 6% Northern European, 10% South West Asian, 19% Sub-Saharan African, and 2% South East Asian - Based on 9 ancestral regions: East Asian, Mediterranean, Southern African, Southwest Asian, Oceanian, Southeast Asian, Northern European, Sub-Saharan African, and Native American.
🔗National Geographic - 404
In 2012, a genetic study focused on North Africa's human populations was conducted. The study highlights the complex genetic makeup of North Africa which showed a genetic composition of harboring a significant local component that became more distinct around 12,000 years ago, possibly influenced by migrations, population expansions, or other demographic events. According to David Comas, coordinator of the study and researcher at the Institute for Evolutionary Biology, "some of the questions we wanted to answer were whether today's inhabitants are direct descendants of the populations with the oldest archaeological remains in the region, dating back fifty thousand years, or whether they are descendants of the Neolithic populations in the Middle East, which introduced agriculture to the region around nine thousand years ago. We also wondered if there had been any genetic exchange between the North African populations and the neighbouring regions and if so, when these took place". The findings reveal a distinct native genetic component in North Africans, setting them apart from Sub-Saharan Africans and aligning them more closely with West-Eurasians, primarily Middle Easterners and Europeans. Though the study emphasizes a dominant genetic lineage in contemporary North Africans tracing back to around 12,000 years ago, it doesn't dismiss the likelihood of genetic continuity from ancient human groups present in North Africa over 60,000 years ago.
🔗Genomic Ancestry of North Africans Supports Back-to-Africa Migrations - PMC (nih.gov)
🔗Population genetics of 17 Y-STR markers in West Libya (Tripoli region) - PubMed (nih.gov)
🔗Ethnic composition and genetic differentiation of the Libyan population: insights on Alu polymorphisms - PubMed (nih.gov)
🔗Genetic ancestry of a Moroccan population as inferred from autosomal STRs - PMC (nih.gov)
🔗HLA Class I and Class II Alleles and Haplotypes Confirm the Berber Origin of the Present Day Tunisian Population - PMC (nih.gov)
The analyses performed showed that current North Africans are closely related to Tunisian (Zrawa and Matmata) and Moroccan (Sousse-Agadir and Eljadida) Berbers, suggesting that North Africans have a genetic Berber profile. On the contrary, North Africans displayed a greater distance from the Arabs of Levant (Palestinians, Syrians, Lebanese, and Jordanians), indicating low genetic contribution of Phoenician and Levant Arab invasion of North Africa. These observations based on HLA markers prompted the conclusion that all Berbers of North Africa constitute a homogeneous genetic unit, except for small isolates, such as the Berbers of Djerba, who display a Berber genetic profile.
🔗https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0192269
A genetic study published in the European Journal of Human Genetics in Nature (2019) showed that Northern Africans are closely related to West Asians as well as Europeans, and Northern Africans can be distinguished from West Africans and other African populations dwelling south of the Sahara.
Research from 2020 on 80 individuals from Southern Tunisia, discovered that Berbers exhibit a mostly indigenous Northwest African genetic make up, characterized by a high amount of native autochthonous North African ancestry. In contrast, the self identified Arab groups had higher Middle Eastern input than the populations in Algeria or Morocco, with some Arab tribes being made-up of assimilated Berbers, or admixed with them.
 |
| *credit:🔗Map of Genetic Admixture of Tunisians from Different Regions by Reddit User CarthageBrigadier |
According to Lucas-Sánchez, Marcel et al. (2021) despite the geneflow from the Middle-East, Europe and sub-Saharan Africa, an autochthonous genetic component that dates back to pre-Holocene times is still present in North African groups. The analysis also showed as a whole no genetic pattern of differentiation between Tamazight (i.e. Berber) and Arabs.
🔗Population history of North Africa based on modern and ancient genomes - PubMed (nih.gov)
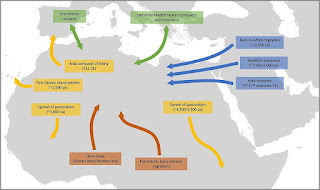 |
| Map Showing the Major Population Movements in Northern Africa |
Pfennig, Aaron et al. (2023), confirmed many previous results, that show many studies are revealing a lack of differentiation between Maghrebi Arabs and Berbers (Imazighen), and that most Arab and Imazighen populations are weakly genetically differentiated.
🔗https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10118306/
A 2024, restated many previous findings such as Amazigh having a high Taforalt component, with this Epipaleolithic ancestry showing genetic continuity that is present in all present North African populations. There were subsequently Neolithic movements from the Middle East and Europe, that diluted the autochthonous Paleolithic component. They also found that there was some differentiation between the Arabs and Amazigh, with the former having a higher Middle-Eastern component.
🔗https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03341-4
Other Relevant Data!
 | |
|
Archive Last Edited: 28/10/2024






Comments
Post a Comment